BECCY-Modelling
Dealing with the process and compilation of the collected data is the main task of our data projects.
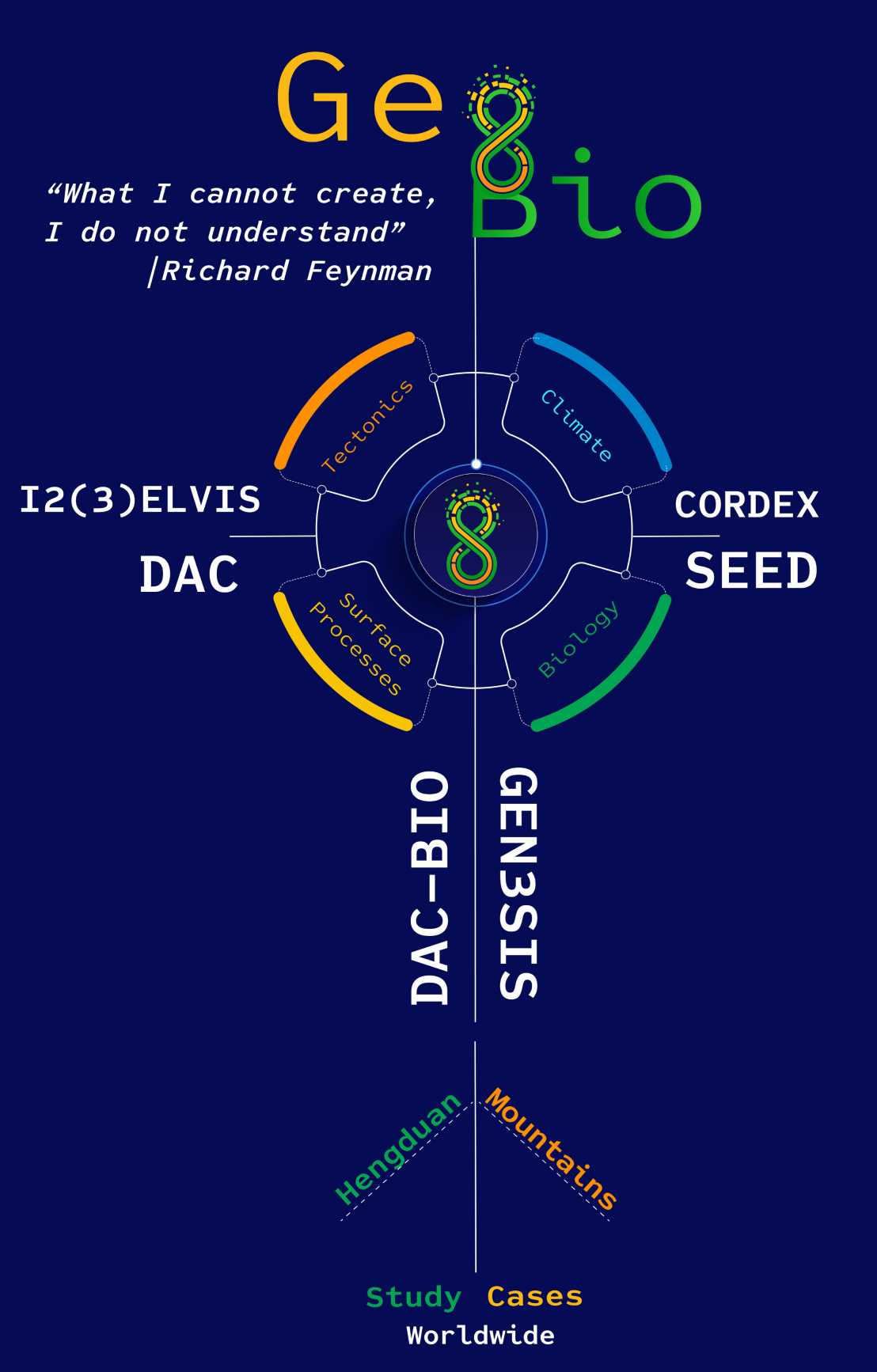
Our approach follows Richard Feynman’s famous words “What I cannot create, I do not understand”, by modelling phenomena and using deduction to understand the geo-bio coupling better. The best available confirmation of the understanding of the causes of a phenomenon is the ability to create a model from first principles and reproduce emergent behaviour and patterns within a system. The modelling principle is as Feynman described it first we guess the theory, then we model the results of our guess, to see if the law behind the guess is correct and then compare it directly with empirical observations to validate it. In this section, we present a summary of the components of the models that we aim to use in testing hypotheses of coupled geo-biology processes.
We have put together a short video to make the complex content as easy to experience as possible.
Landscape Evolution Models (LEMs) solve physical laws of surface erosion, sediment transport, and sediment deposition as a function of tectonic and climatic forcing operating on a numerical grid representing the surface of the Earth. The core of LEMs is an algorithm for routing water across the landscape through a channel network and the incision of these channels into bedrock.
Prof. Dr. Sean D. Willett and his collaborators have worked extensively on LEM development and have presented two important solutions to the multi-scale resolution problem. The first was just to refine the grid down to the meter scale to resolve discrete features. This was possible through the construction of an innovative structured stack algorithm, that exploited the tree structure of the channel network to reduce the dimensionality of the numerical model, thus making possible solutions with millions of nodes. In an alternative model, he augmented it with an algorithm in which analytical solutions for divide position were added to the numerical model. Together with an adaptive, irregular grid, this model is capable of exact positioning of discrete features such as drainage divides, using a fairly coarse grid to define the major channel network, and adding in the finer resolution through analytical, sub-grid solutions.
This latter model is called DAC (Divide and Capture) based on irregular and dynamic grid because tectonic motions are easily implemented with an irregular grid by moving nodes with the tectonic velocity and recalculating a Delauney tessellation. This avoids the numerical dispersion caused by a fixed grid method and preserves the topology of the river network. A number of important applications have been demonstrated, including shear tectonics in New Zealand, drainage divide migration and collisional tectonics in the Three Rivers Region of the Hengduan Mountains. The code has also been coupled to the 3-dimensional geodynamic code of Prof. Dr. Taras Gerya demonstrating the viability to model large-scale, 3-dimensional problems.
Prof. Dr. Taras Gerya and his collaborators have developed a family of 2D and 3D geodynamic modelling codes known as I2(3)ELVIS that allows for high-resolution thermomechanical modelling of various plate tectonic processes. These processes range continental breakup and ridge-transform spreading patterns to subduction zones and continental collision processes. The numerical approach is based on staggered finite differences and marker in cell techniques that demonstrated their numerical stability and low computational costs that are essential for running high-resolution 4D (space-time) thermomechanical numerical. The numerical approach accounts for non-linear viscoplastic rock rheology based on experimentally calibrated flow laws and allows for large deformation, spontaneous fault development and accurate surface topography treatment This approach has also been successfully applied to complex geometries of continental corner collision that is highly relevant for the Himalayan-Hengduan region.
Climate models are based on the same fundamental equations as the atmospheric models, but also include dynamical representations of oceans and sea-ice, and play a key role in providing projections of anthropogenic climate change. Prof. Dr. Christoph Schär and his collaborators have used the increase in computing power and the last decade, to foster a large improvement of the resolution of RCMs, which can serve a wide range of purposes. Thus, he created a set of CORDEX simulations through the European Copernicus Climate Change Services program. The respective RCM simulations follow the internationally coordinated protocol in terms of domain, resolution, scenario, and simulation period. Simulations are also made for South Asia and consider simulations over the East Asian domain relevant for the current project.
A goal is fostering the potential of using a regional climate model to investigate the past climate at a resolution that is relevant to describe the ecological conditions determining species distribution in complex mountain systems. High resolution climate modelling will allow to capture ecological gradients associated to biodiversity at high spatial resolution.
How species come to exist, how they evolve, how they interact, how they go extinct within shifting environmental boundaries is key to understanding the shaping of biodiversity ecosystem processes. Simulation models of biodiversity have principally focused on implementing general formulations for biological processes including speciation, extinction, ecological interactions and phenotypic evolution. Simulations of the dynamics biodiversity in space and time focusing on multiple taxonomic groups are expected to give information about processes shaping biodiversity organization and evolution. As a side effect, empirical biological patterns should provide information about different aspects of the geological and climatic history of a region.
Prof. Dr. Loïc Pellissier and his collaborators have developed a process-based simulation model of biodiversity, called Speciation-Evolution-Extinction-Dispersion (SEED) Models. The development of SEED models requires scenarios about the dynamics of the underlying geography, in particular the movement of tectonic plates and orogenic processes and the relationship between that geography and climate. Together with Prof. Dr. Niklaus Zimmerman, develops and evaluates empirical and mechanistic models the ecological, functional and phylogenetic drivers and mechanisms that shape biodiversity and community compositions across spatial and temporal scales. The evaluation of numerical models can be achieved via the comparison of emergent patterns from models, constrained by biological and landscape processes, to patterns observed in independent data.
The main objective of this project with all the case studies included is to develop a fully coupled model, that both BECCY leaders Prof. Dr. Sean D. Willett and Prof. Dr. Loïc Pellissier are working on actively. The processes of tectonics are associated with changes in local climate, topography complexity and in erosion patterns. Together these changes set the template on which life evolves. The study of these forcings and feedbacks with their spatial and temporal complexities requires a fully integrated process-based model based on solid physical rules.
DAC-BIO is a coupled LEM-biological model and in our current version speciation occurs exclusively through vicariance (allopatric speciation). This new code can model biogeographic evolution of aquatic organisms. In this type of simulations, a landscape in equilibrium with steady, uniform rock uplift rate and uniform rock material properties is subjected to a horizontal velocity simulating wedge accretionn. Horizontal advection moves grid nodes from the lower boundary toward the main divide that separates the pro-wedge side of the orogenic wedge from the retro-wedge side. As nodes are pushed toward the divide, they are captured by rivers. Each river capture results in a pro-wedge population moving to a retro-wedge basin and dispersing throughout the catchment, enhancing species richness. Each capture leads to the creation of a new species, based on the genetic drift that occurs between captures. Species richness therefore reflects the number of river captures each catchment has accrued. The result is a prediction that the advection of the topography results in greater species richness on the retro-wedge. It also shows variance between catchments on the pro-wedge due to lateral captures between basins. The former postdoctoral researcher in the Earth Surface Dynamics Group, Dr. Helen Beeson, works and develops this modelling techniques, with a special focus on aquatic organisms.
Gen3sis is a biodiversity modelling is largely focused on contemporary ecological constrains on species diversity, while modelling the processes of diversification itself in a spatial context remains challenging. Traditional correlative approaches in macroecology may not provide a mechanistic understanding of the way historical processes have shaped the distribution of biodiversity. To disentangle causalities for emergent patterns, the application of dynamic simulation models may provide a better mechanistic understanding on biodiversity gradients.
For that, we are developing GEN3SIS, which is a process-based simulation model and infers biodiversity patterns from historical range dynamics taking into account eco-evolutionary feedbacks. This spatial diversification model will provide an important tool for null hypothesis testing and better understanding of the processes of speciation and extinction, which will be contrasted with empirical data. Case studies will include the diversification of tree assemblages (e.g. within the Fagaceae family), marine system (e.g. shallow reef habitat in interaction with another project specific to coral reef system) and alpine plant assemblages.
Ultimately, the model will inform on the ecosystem responses to global changes, and especially the loss of diversification potential for biodiversity. This may challenge current conservation practice and move current concepts toward an increased focus on the conservation of evolutionary potential.
